Authors: Wako Bungula and Isabel Darcy
Full paper can be found at https://arxiv.org/abs/2409.17360.
Given a topological space equipped with a continuous function , a filtration of covers of gives rise to a filtration of covers of , which in turn gives rise to a filtration of the Mapper graphs. This filtration of Mapper graphs induces a filtration of homology groups. This paper is instead interested in point cloud data.
Filtrations via Nerves of a Cover
Def
Given a family of covers, equipped with a family of maps for some parameter where if then , we say there is a filtration of covers if .
Let be a topological space with coverings, and is the family of functions
where and for , implies that and . Then induces a filtration of simplicial maps
where is a simplicial map defined on the vertex set of such that if then where .
Theorem 1
A filtration of covers induces a well-defined filtration of simplicial complexes.
Theorem 2
A filtration of simplicial complexes induces a filtration of homology groups.
Filtration of Mapper Graphs
The traditional Mapper depends on several parameters such as bin size and percent overlap. Recall that in the Mapper we make use of a filter function and form a cover of that is then pulled back by to create a cover of . Then each pullback cover is clustered to form the cluster cover.
While a filtration of covers of induces a filtration of pullback covers of , it is not clear that it induces a filtration of cluster covers. The paper claims there are instances in which we do not get a filtration of cluster covers. Clearly a filtration of cluster covers induces a filtration of their nerves and hence their homology groups. Thus, we simply only need to check when a filtration of covers induces a filtration of cluster covers.

Remark: This is all for the filtration as defined above (i.e., the Multiscale mapper). We can still construct a filtration starting with the cluster covers (see Steinhaus Filtration and Stable Paths in the Mapper).
No Filtration of Cluster Covers: Complete/Average-Linkage, bin size.
Two common clustering algorithms used in Mapper is single-linkage and DBSCAN as they provide the correct connected components of a dataset. But, two points far from each other may be put in the same cover if there is a chain of points connecting them.
The authors construct an example in which the Mapper parameterized by bin size fails to produce a filtration. More specifically, they take a data set and the filter function and construct two covers of :
- Two interval and with 20% overlap
- Two intervals and with 50% overlap Note that and .
This forms a simple filtration on the dataset
They then used single-linkage clustering on the cover. After clustering, there is a cover that lies in but is not contained any clusters of . In other words, a filtration on the pullback cover parameterized by bin size failed to induce a filtration on the cluster cover.
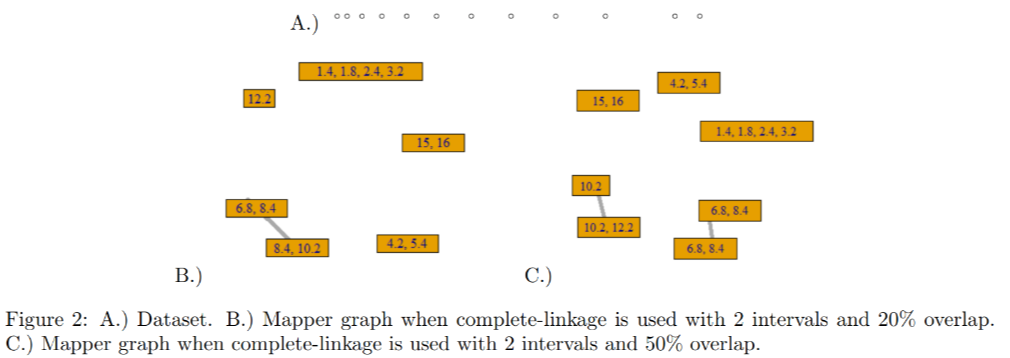
Notice that the cover in the Mapper graph corresponding to (B) is lost in the Mapper graph corresponding to (C).
DBSCAN
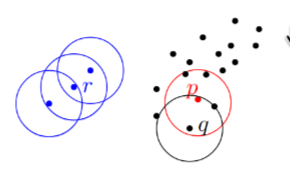
The general idea of DBSCAN is to cluster a set of dense points together, and if there is a set of low-density points they are considered noise. DBSCAN is configured by two parameters: and radius . An -neighborhood of a point is considered a dense set if the neighborhood contains at least points, i.e.,
KeplerMapper defaults to and .
- A point is a core point if
- A point is a border point if and there is a core point such that .
- A point is noise if is neither a core point nor a border point.
Def
A point is directly density-reachable from a point w.r.t. and if is a core point and .
Def
A point is density-reachable from a point w.r.t. and if there is a sequence of points such that is directly density-reachable from .
Def
A point is density-connected to a point w.r.t. and if there is a point such that both and are density reachable from w.r.t. and .
Question: Why the distinction between density-reachable and density-connected? It seems like being density-reachable from implies that and are density-connected.
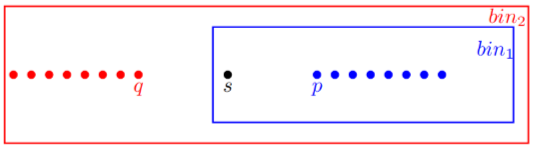
Answer: Density-reachable is not a symmetric relation. For example, in the dataset below

the point is (directly) density-reachable to the points and , but and are not density-reachable to because is not a core point (). But is density-connected to because there is a point such that and are density-reachable from .
Def
Let a dataset where is a set of noise points and is a cluster w.r.t. and satisfying:
- (Maximality) , if , , and is density-reachable from w.r.t. and , then .
- (Connectivity) , is density-connected to w.r.t. and .
In summary, a DBSCAN cluster of a dataset is a set of points such that any two points are density-connected and there is no larger such cover .
The paper constructs an example in which the assignment of a point is dependent on the order the data is listed.
Def
A point is a free-border point w.r.t. and if there exists two points and such that
- and
- and
- , and
- is not density connected to .
For the example, the dataset given below:

contains a free-border point when and .
The paper claims that as long as there are no free-border points a filtration of covers of gives a filtration of cluster covers of as bin size increases, the parameter increases, or decreases.
Lemma 2
Let be a dataset.
- If is a core point, then there is a cluster containing and if then is density-reachable from .
- Let be a cluster. Then there is a point that is a core point. If then is density-reachable from .
In other words, the Lemma above states that a cluster is determined by any of its core points and the core points do not change depending on the order of the dataset.
Filtration of cluster covers: DBSCAN, bin size.
Free-border points can results in a failure to filter the cluster covers. Consider the example below:
Assume that is ordered before and , and . When DBSCAN is applied to a cluster is formed w.r.t. and containing and all points to the right of . When we apply DBSCAN to we get two clusters w.r.t. and : contaning points , and points to the left of and containing and points to the right of .
This is an issue as but . Thus, the free-border point results in a filtration of cluster covers parameterized by bin size being invalid.
The critical point here is the free-border point . In the absence of free-border points the issue disappears:
Lemma 4
Suppose there are no free-border points when DBSCAN is used to cluster. If ., then .
Therefore, if there are no free-border points than there is a filtration of cluster covers parameterized by bin size. As a corollary, if then there is a filtration of cluster covers parameterized by bin size as there will be no free-border poitns.
Filtration of cluster covers: DBSCAN, ϵ
Notice that if then if is a core point w.r.t then is a core point w.r.t. . Thus, one could potentially construct a filtration parameterized by .
Lemma 5
Suppose is a data set, , and and are fixed. If there are no free-border points w.r.t. and then .
Therefore, the absence of free-border points indicates that there is a filtration of cluster covers parameterized by .
Filtration of cluster covers: DBSCAN, MinPts
Notice that if and is a core point w.r.t. and , then is also a core point w.r.t. and .
Lemma 6
Suppose is a dataset, , and and are fixed. If there are no free-border points w.r.t. and then .
Therefore, the absence of free-border points indicates that there is a filtration of cluster covers parameterized by decreasing .
Filtration of Simplicial Complexes and Homology Groups
If there are no free-border points, then we can construct simplicial and homological filtrations parameterized by
- Bin size
Bi-Filtrations and Stability
The paper claims that DBSCAN is not stable under small perturbation. Let be a dataset and the dataset obtained by perturbing by at most , i.e., there is some function such that for all . We say that
How does applying DBSCAN to and vary?
For example, consider the following two datasets:
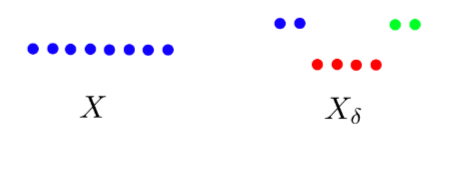
If we let be the distance between two points in and , then is clustered into a single set where as is clustered into three disjoint sets.
We construct a two-dimensional filtration (bi-filtration) parameterized by and bins , leaving fixed. Note that each parameter alone gives a filtration (assuming no free-border points), that is, given a dataset there is a filtration of cluster covers
which induces a filtration of simplicial complexes
which induces a filtration of -th homology groups
A similar set of filtrations exist parameterized by . Combining the two together gives a set of bi-filtrations where is one dimension and is the other.
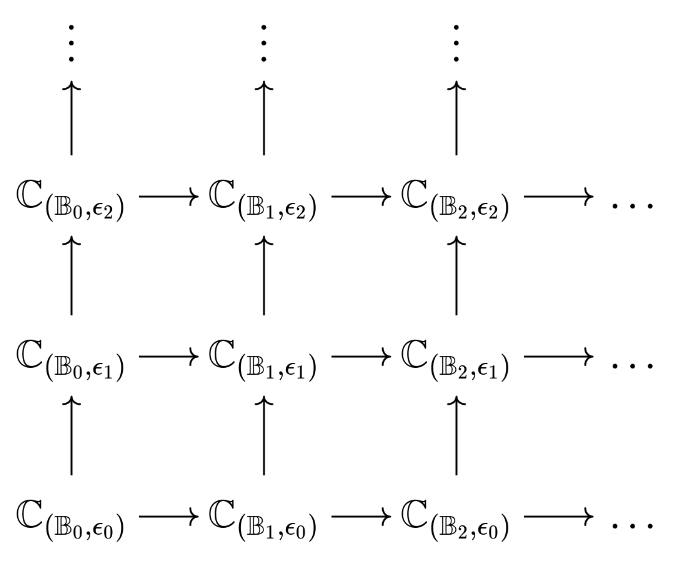
We can perform the same with to get another set of bi-filtrations .
Interleaving of Bi-filtrations
See The structure and stability of persistence modules for more details.
Def
Let be a polynomial ring in variables . An -graded module is a module such that and for all , where is a vector space over some field . The action of gives rise to a linear map for all .
This is unnecessarily complicated. We’re just taking persistence modules parameterized by two variables. Thus, we have a set of vector spaces and a set of linear maps for all .
Def
For an -graded module and , is the shifted module such that .
Def
For and -graded module, and ,
is the (diagonal) -transition morphism such that .
This is just -homomorphisms but now .
Def
Let . Two -modules and are -interleavd if there are morphisms and such that and .
Again, this is just -interleaving but now .
Stability Against Perturbation
Assume there are no free-border points. Then as described above we get a filtration for
and a filtration for
According to proposition 1, there are family of maps and such that the diagram below commutes:
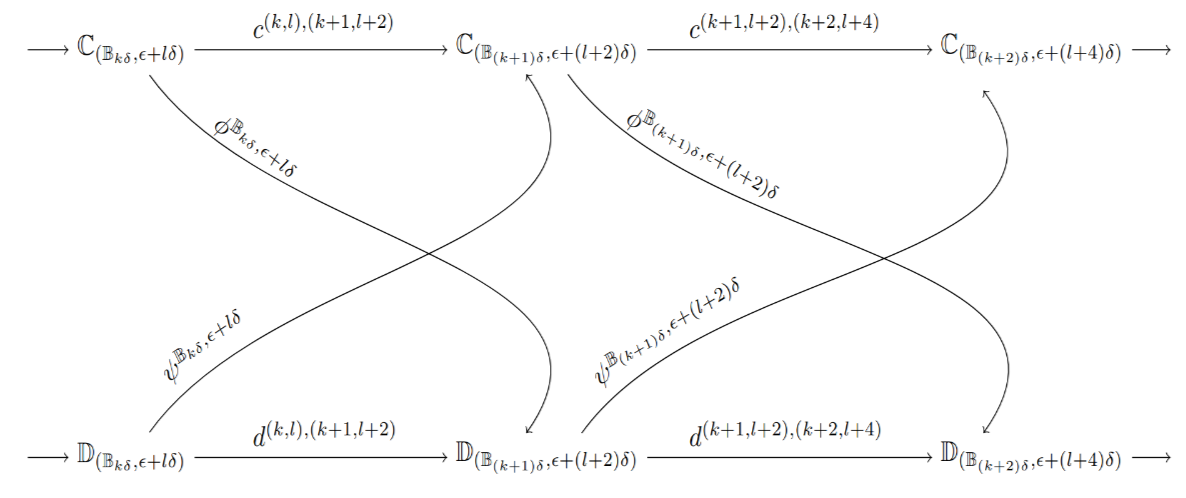
In other words, the filtrations and are -interleaved. This -interleaving induces a -interleaving between their homologies and .